Beyond Black-Box Models: Advances in Data-Driven ADMET Modeling
AI models hope to provide accurate predictions of ADMET properties, which remain a major bottleneck in drug discovery.
Complete the form below to unlock access to ALL audio articles.
Predicting how a drug candidate behaves in the body, including its absorption, distribution, metabolism, excretion and potential toxicity (ADMET), is a cornerstone of modern drug discovery. Despite decades of development, both experimental and computational methods struggle with inconsistent data quality, species-specific bias and high regulatory expectations. While open-source AI ADMET prediction models have shown significant progress in recent years, most of them still struggle with limited interpretability, inflexibility and a lack of validation. Here, we describe how Receptor.AI’s ADMET prediction approach effectively addresses these limitations through a combination of multi-task deep learning methodologies, graph-based molecular embeddings and rigorous expert-driven validation processes. A comprehensive preprint describing Receptor.AI’s ADMET model, including its architecture and validation methodology, is now available on bioRxiv 1.
Modern challenges
Regulatory agencies such as the US Food and Drug Administration (FDA) and European Medicines Agency (EMA) require comprehensive ADMET evaluation of drug candidates to reduce the risk of late-stage failure. Both agencies operate under distinct but globally influential regulatory frameworks, shaping drug development standards worldwide. For example, CYP450 inhibition and induction studies are essential for assessing potential metabolic interactions, while hERG assays remain a cornerstone in identifying cardiotoxicity risks.2,3 Liver safety evaluation is also a critical part of early drug screening, as hepatotoxicity has been a common factor in post-approval drug withdrawals.4,5
The FDA and EMA recognize the potential of AI in ADMET prediction, as long as models are transparent and well-validated.6,7 Yet significant challenges persist, particularly around inconsistent data quality and a lack of standardization across heterogeneous ADMET datasets, which undermine models’ reproducibility and limit their generalization. While the FDA mandates comprehensive in vitro and in vivo data for assessing drug–drug interactions and transporter effects, AI can assist in prioritizing endpoints and selecting compounds in the preclinical stages. For AI adoption to gain traction, issues around black-box nature, dataset bias, and generalization to novel chemical structures must be addressed through standardized validation protocols and benchmarks. Rather than replacing traditional ADMET evaluations, AI serves as an additional predictive layer to streamline regulatory submissions, decrease experimental burden and optimize the drug selection process. Validated AI tools can support compliance, strengthen safety assessments and build regulatory trust.
On April 10, 2025, the FDA took a major step by outlining a plan to phase out animal testing requirements in certain cases.8 The roadmap formally includes AI-based toxicity models and human organoid assays under its New Approach Methodologies (NAMs) framework. These tools may now be used in Investigational New Drug and Biologics License Application submissions, provided they meet scientific and validation standards. The plan includes pilot programs and defined qualification steps to guide adoption.
The limitations of traditional ADMET assessment approaches
Traditional ADMET assessment methods remain central to early drug development but are difficult to scale. In vitro assays, such as cell-based permeability and metabolic stability studies, along with in vivo animal models, are slow, resource-intensive and not designed for high-throughput workflows (Figure 1). As compound libraries grow, these methods become increasingly impractical.9 Early computational approaches, especially quantitative structure-activity relationship (QSAR)-based models, brought automation to the field using predefined molecular descriptors and statistical relationships.10 However, their static features and narrow scope limit scalability and reduce performance on novel diverse compounds, especially as discovery efforts demand broader coverage of the chemical space and faster turnaround.
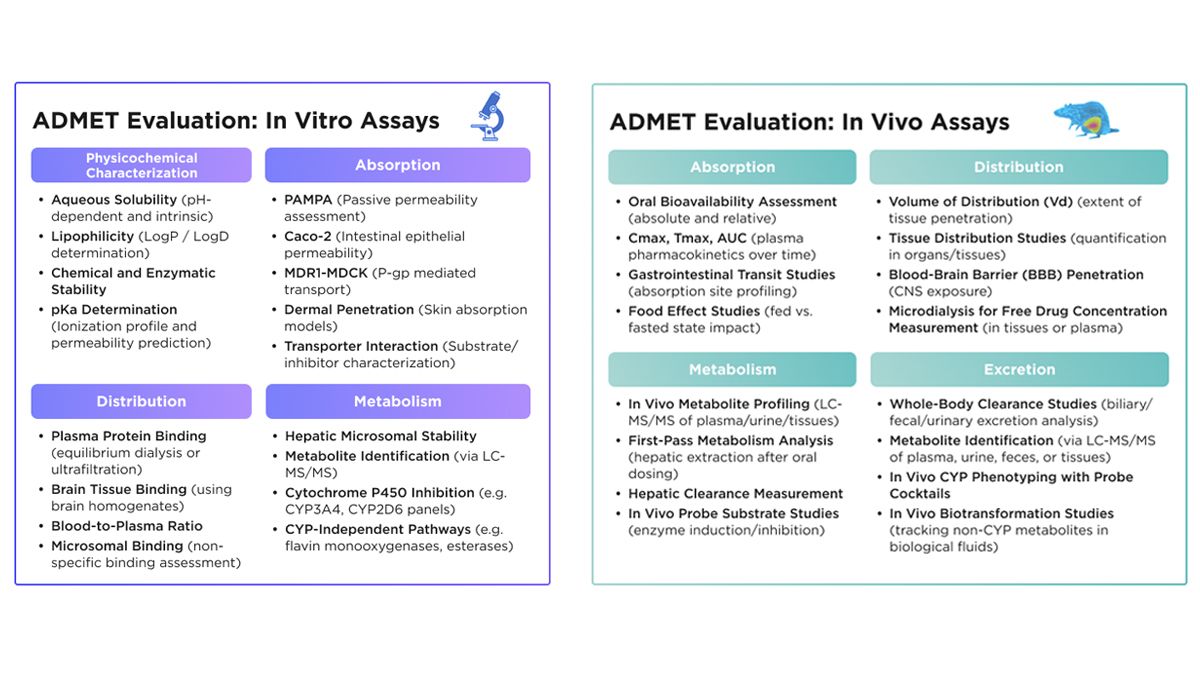
Figure 1: Overview of the standard in vitro and in vivo ADME assays used to evaluate a drug’s absorption, distribution, metabolism and excretion properties during preclinical development. Credit: Receptor.AI
Tools such as pkCSM and ADMETlab are AI-based predictive models that have contributed meaningfully to ADMET property estimation by offering user-friendly platforms for toxicity and pharmacokinetic endpoint prediction. ADMETlab 3.0 is actively maintained and now incorporates partial multi-task learning to improve the prediction of related endpoints.11 Despite this progress, these tools still frequently implement simplified representations and single-task or limited multi-task machine learning (ML) frameworks that may not fully capture the complex interdependencies among pharmacokinetic and toxicological endpoints (Figure 2). While frequently used as benchmarks in comparative studies, their underlying architectures differ substantially from newer deep learning models built for dynamic, multi-endpoint prediction and integration of high-dimensional chemical data.
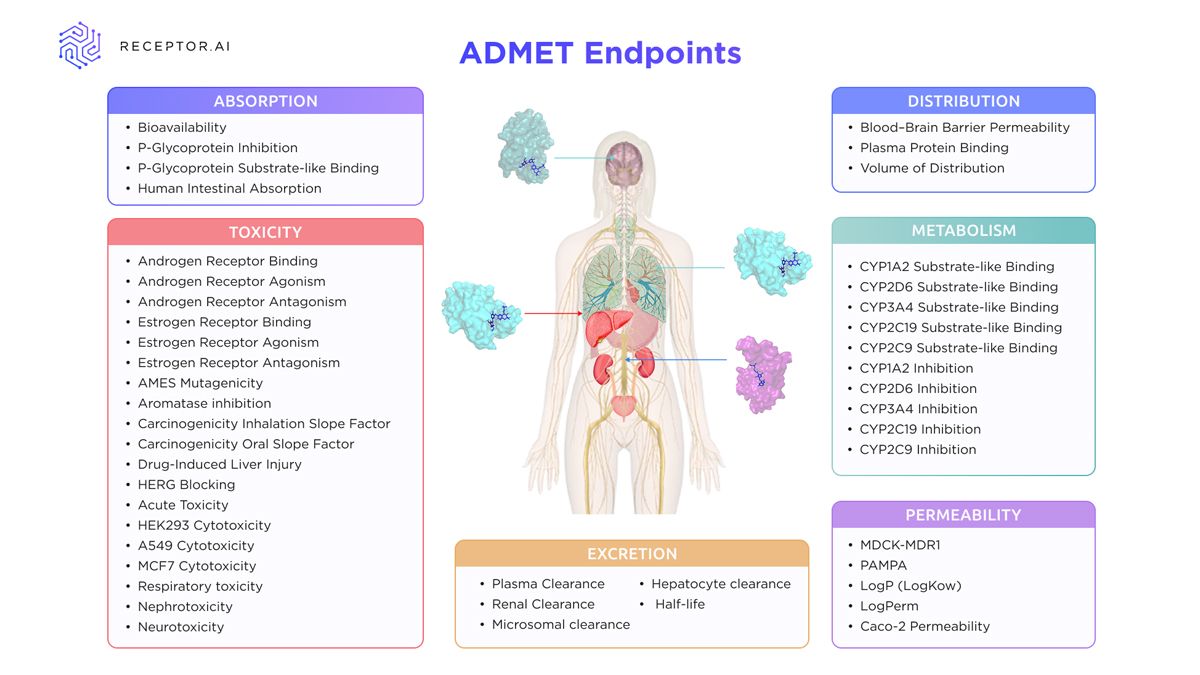
Figure 2: Overview of ADMET endpoints supported by Receptor.AI, covering key parameters across absorption, distribution, metabolism, excretion, toxicity and permeability relevant to early-stage drug development. Credit: Receptor.AI
Moreover, most AI-based models in drug discovery function as black boxes, generating predictions without clear attribution to specific input features. This opacity stems from the complexity of deep neural network architectures, which obscure the internal logic driving their outputs. In ADMET modeling, such a lack of interpretability hinders scientific validation and regulatory acceptance, where clear insight and reproducibility are essential.
The species-specific metabolic differences add another layer of complexity, which can mask human-relevant toxicities and distort the results for other endpoints. Historical cases like thalidomide and fialuridine underscore the limitations of traditional preclinical testing in capturing human risks. As black-box AI tools proliferate, their limited transparency has become a regulatory concern. Overcoming these barriers requires a new generation of ADMET models built on multi-task learning, continuous adaptation to evolving datasets and transparent architectures that can support both scientific discovery and regulatory trust.
Limitations of current open-source ADMET models
Open-source ADMET models have gained traction in drug discovery due to their accessibility and ease of integration. However, they still face fundamental limitations that restrict their real-world effectiveness. Many rely heavily on QSAR methodologies and static molecular descriptors, limiting their ability to accurately represent complex biological interactions.12 These models typically utilize simplified 2D molecular representations, lack adaptability to new data and struggle to generalize predictions for structurally diverse compounds. Additionally, their dependence on narrow or outdated datasets further reduces predictive robustness, particularly in regulatory and translational settings where both accuracy and coverage are critical.
New open-source ADMET platforms contributed meaningful advances in molecular property prediction, yet key limitations persist. Chemprop employs message-passing neural networks and performs well in multitask settings, but its latent representations are not easily interpretable at the substructure level.13 DeepMol enables rapid prototyping through AutoML-based workflows, though its abstraction layers reduce transparency and model explainability.14 OpenADMET is developing a community-driven ADMET resource, but lacks large-scale benchmarking and draws from fragmented, non-standardized datasets.9 ADMET-AI combines Chemprop with RDKit to enhance prediction quality, yet its static architecture does not support species-specific modeling or online adaptation.15
Receptor.AI ADMET prediction model
As outlined earlier, current ADMET prediction models often struggle with key limitations: many rely on shallow feature extraction, show bias toward specific organisms or chemical classes and lack the architectural flexibility needed for refinement. These constraints limit both their performance and applicability in diverse, real-world drug discovery scenarios.
To overcome these challenges, Receptor.AI’s ADMET model consists of two interconnected parts: Mol2Vec (inspired by the Word2Vec language encoder) encodes molecular substructures into high-dimensional vectors, which are then combined with selected chemical descriptors and processed through multilayer perceptrons to predict 38 human-specific ADMET endpoints (Figure 3). This design enables the model to capture complex, structure-driven relationships relevant to pharmacokinetics and toxicity. The architecture is inherently flexible, allowing users to fine-tune existing endpoints on new datasets or train custom endpoints tailored to specific research needs. We evaluate not only individual predictions, but also how endpoints interact, scoring their correlations, dependencies and cross-predictive patterns. A separate LLM-based rescoring is used to predict a final consensus score for each compound by integrating signals across all ADMET endpoints. This enables the model to capture broader interdependencies that simpler systems often miss, improving consistency and predictive reliability.
The model is available in four variants, each reflecting a distinct strategy for descriptor integration and optimized for different virtual screening contexts in terms of predictive accuracy and computational efficiency. Mol2Vec-only is the fastest model that relies solely on substructure embeddings derived from Morgan fingerprints. Mol2Vec+PhysChem adds basic physicochemical properties such as molecular weight, logP and polar surface area. Mol2Vec+Mordred incorporates a comprehensive set of 2D descriptors from the Mordred library, offering a broader chemical context. The most advanced version, Mol2Vec+Best, is the most accurate but also the slowest variation. It combines Mol2Vec embeddings with a curated set of high-performing molecular descriptors selected through statistical filtering. This version is the subject of the recently released preprint 1. All model variants support CSV and SDF input formats, implement SMILES standardization and apply feature normalization to ensure consistency across datasets.
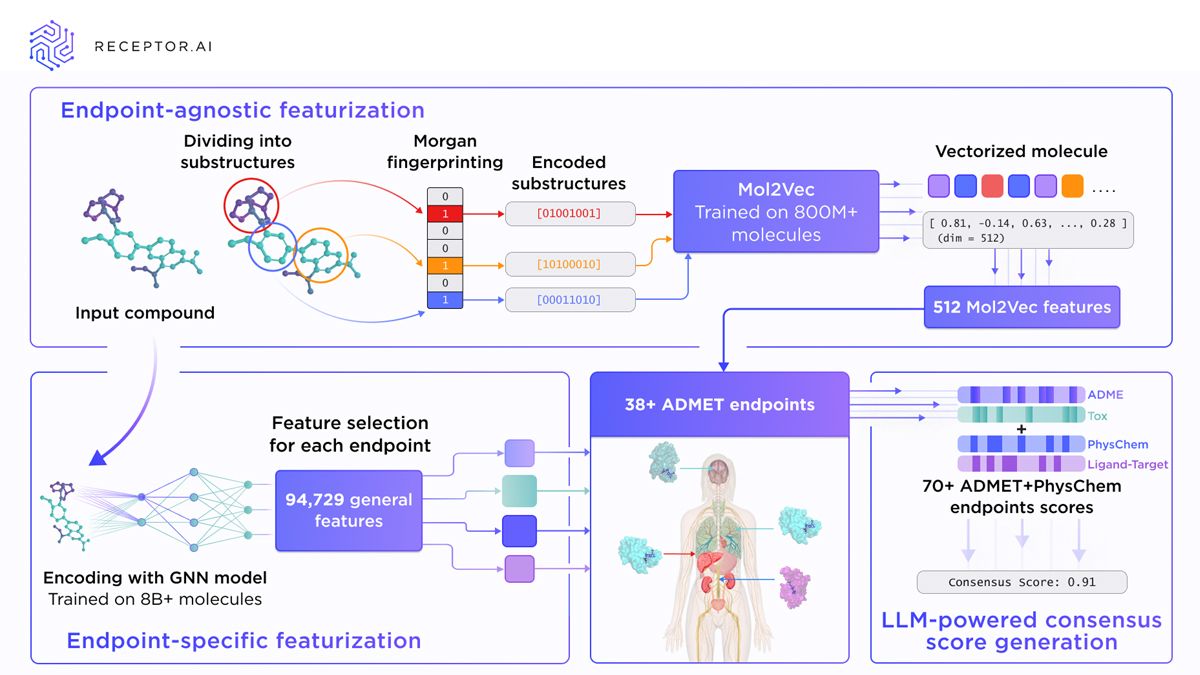
Figure 3: Overview of Receptor.AI’s ADMET prediction workflow, combining endpoint-agnostic and endpoint-specific molecular featurization with LLM-assisted consensus scoring across over 70 ADMET and physicochemical endpoints. Credit: Receptor.AI
Conclusions
Accurate prediction of ADMET properties remains a major bottleneck in drug discovery, constrained by sparse experimental data, interspecies variability and evolving regulatory requirements. Open-source AI models have improved baseline performance, but many continue to face limitations in interpretability, adaptability to novel chemical space and the robustness of their validation frameworks. The ADMET model developed at Receptor.AI addresses these challenges by integrating multi-task learning, graph-based molecular embeddings and curated descriptor selection.





